|
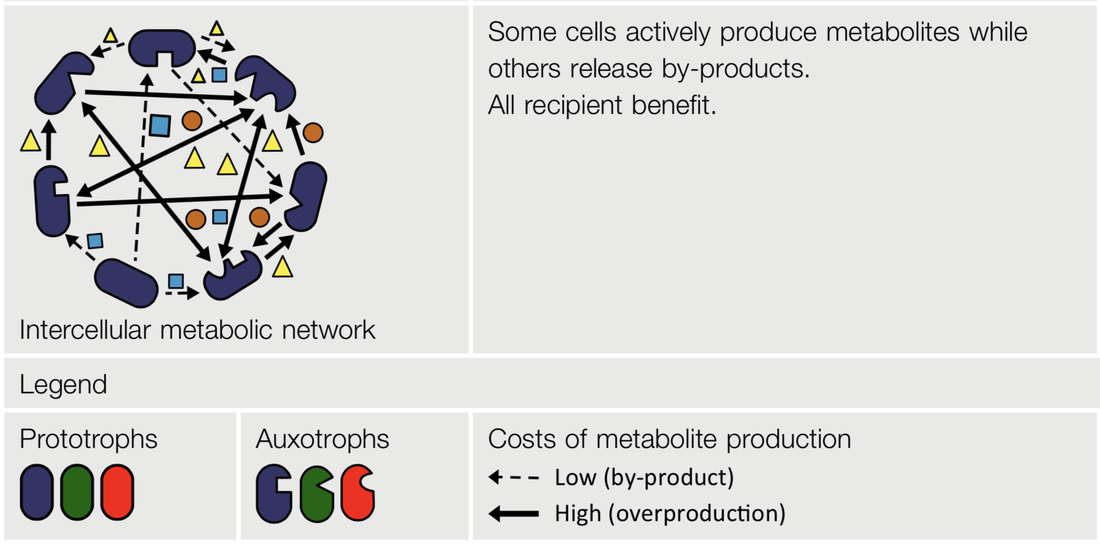
The frequent loss of metabolic genes from microbial genomes along with intricate patterns of cross-feeding, where other coexisting genotypes compensate the functional deficiency, suggests that within bacterial consortia, metabolism is often a community-level property and not a feature of an individual cell anymore. If true, this implies that a bacterial communities' metabolism is in essence a super-metabolic network, where many of its constituents perform specialized biosynthetic or catabolic tasks that also benefit other members of the community.
|
Indeed, recent metagenomic and empirical surveys of environmental bacterial communities have revealed that individual genotypes in a community can have distinct patterns of amino acid auxotrophies, such that some members lack multiple biosynthesis pathways, and, at the same time, specialize in the production of another set of amino acids.
The establishment of such interactions is driven by a complex interplay between fitness-advantages of individual mutants and the eco-evolutionary dynamics between multiple genotypes. Ultimately, the highly-conserved structure of core metabolic pathways in otherwise rather divergent bacterial lineages could guide the evolutionary self-organization of metabolic exchange even between very different bacterial species.
In this project we combine bacterial community data from soil samples with theoretical models to fully understand the ecology and evolutionary dynamics of intercellular metabolic networks.
The establishment of such interactions is driven by a complex interplay between fitness-advantages of individual mutants and the eco-evolutionary dynamics between multiple genotypes. Ultimately, the highly-conserved structure of core metabolic pathways in otherwise rather divergent bacterial lineages could guide the evolutionary self-organization of metabolic exchange even between very different bacterial species.
In this project we combine bacterial community data from soil samples with theoretical models to fully understand the ecology and evolutionary dynamics of intercellular metabolic networks.
Researchers:
Leonardo Oña, Shryli Kedambadi Shreekar, Christian Kost
Selected Publications:
Pande S, Kost C. (2017). Bacterial unculturability and the formation of intercellular metabolic networks. Trends in Microbiology, 25(5), 349-361. doi:10.1016/j.tim.2017.02.015
Oña L, Kost C. (2022) Cooperation increases robustness to ecological disturbance in microbial cross-feeding networks. Ecology Letters. https://doi: 10.1111/ele.14006.
Oña L, Kost C. (2022) Cooperation increases robustness to ecological disturbance in microbial cross-feeding networks. Ecology Letters. https://doi: 10.1111/ele.14006.